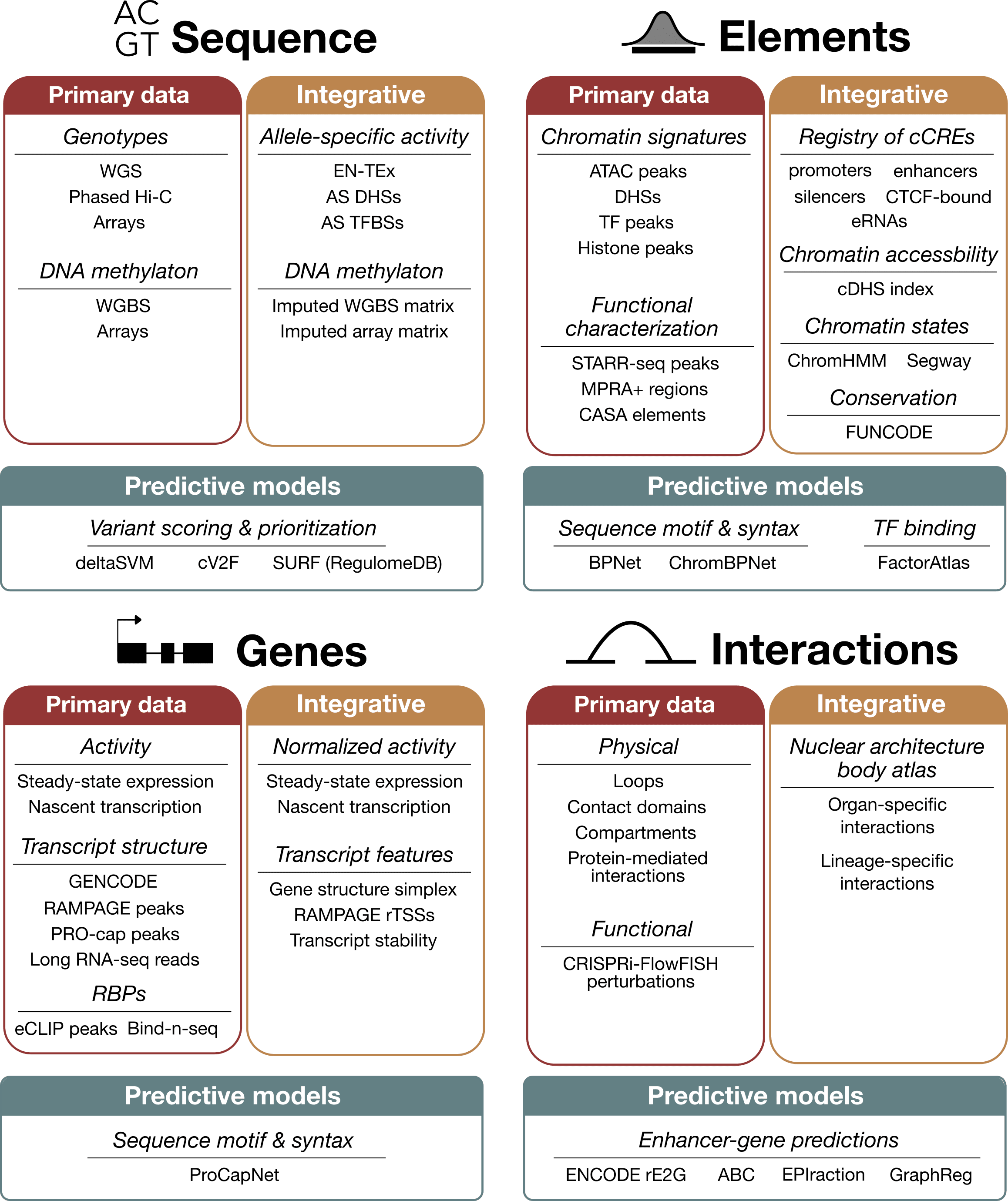
About SCREEN
SCREEN (Search Candidate Regulatory Elements by ENCODE) is a web-based visualization and discovery platform for exploring the ENCODE Registry of cis-Regulatory Elements (cCREs) and other ENCODE Encyclopedia annotations. SCREEN enables users to query, visualize, and download regulatory annotations across hundreds of human and mouse cell and tissue types and to examine their relationships to genes, transcription factor binding, chromatin state, and three-dimensional genome organization.
The ENCODE Encyclopedia
The ENCODE Encyclopedia is an integrated collection of functional genomic annotations derived from large-scale experimental data generated by the ENCODE Project. These annotations span multiple conceptual layers, including DNA sequence features, regulatory elements, genes and transcripts, and chromatin interactions. Some annotations derive directly from primary experimental data, while others reflect integrative analyses that combine multiple assays to provide higher-level functional interpretations.
SCREEN provides an interactive interface for navigating these annotations, centering many views on cCREs as a unifying framework for regulatory analysis.
The Registry of Candidate cis-Regulatory Elements (cCREs)
The Registry of cCREs is a genome-wide catalog of candidate regulatory elements defined using chromatin accessibility, histone modification, and transcription factor binding data generated by ENCODE. The Registry is designed to be comprehensive, cell type–agnostic, and extensible as new data become available
Element Anchors
In ENCODE4, cCREs are anchored using multiple complementary strategies to capture a broad spectrum of regulatory elements:
- Representative DNase hypersensitive sites (rDHSs), generated by clustering DNase-seq peaks across large numbers of biosamples, define the majority of accessible regulatory regions.
- Transcription factor clusters, representing reproducible transcription factor binding sites that do not overlap DNase hypersensitive regions, capture regulatory elements with low chromatin accessibility but strong, protein-binding signatures.
- Multi-mapper rDHSs, derived from alignments that retain multi-mapping reads, recover regulatory elements in recently duplicated or repetitive genomic regions that were underrepresented in earlier Registry versions.
Together, these anchors define millions of candidate regulatory elements in the human and mouse genomes.
Quantifying Epigenomic Signals
For each cCRE anchor, we compute standardized signal summaries for chromatin accessibility (DNase-seq or ATAC-seq), histone modifications (H3K4me3 and H3K27ac), and CTCF binding across all available biosamples. Signals are converted to Z-scores within each biosample to ensure comparability across assays and experiments. These standardized signals form the basis for both classification and biosample-specific activity annotations.
cCREs defined using multi-mapping–inclusive chromatin accessibility data do not have associated epigenomic signal tracks, as the full set of ENCODE assays was not reprocessed using multi-mapping–inclusive alignments. These elements are therefore included in the Registry as candidate regulatory elements but lack quantitative signal annotations.
Cell Type–Agnostic Classification of cCREs
Each cCRE is assigned a cell type–agnostic class based on its dominant biochemical signatures across all surveyed biosamples and its proximity to annotated transcription start sites (TSSs). This classification is intended to be analogous to gene catalogs, which define genes independently of their expression levels in individual cell types.
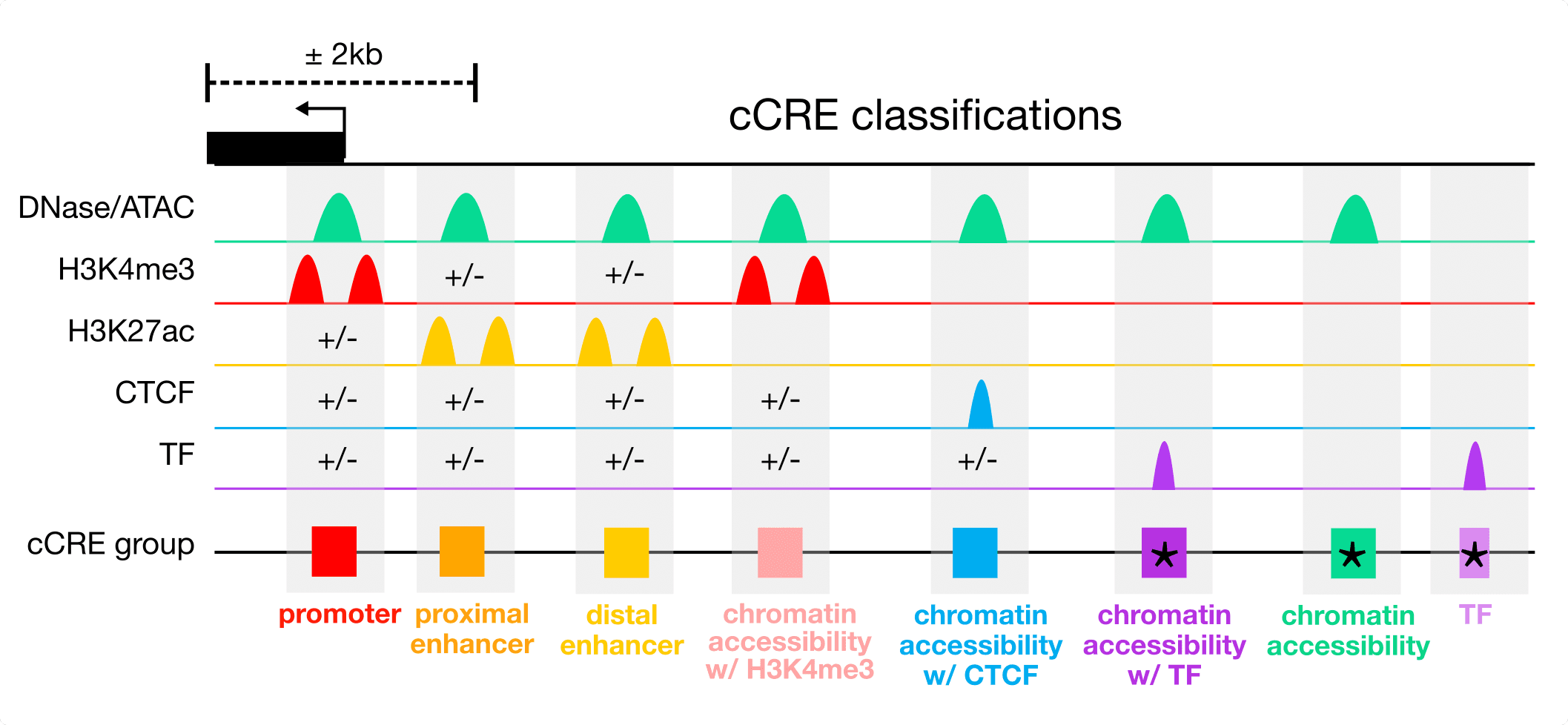
The major cCRE classes include:
Enhancer-like cCREs: Accessible elements with high H3K27ac signal, subdivided into TSS-proximal and TSS-distal enhancers based on distance to the nearest TSS.
Chromatin accessibility + H3K4me3 (CA–H3K4me3) cCREs: Accessible elements with H3K4me3 but lacking strong H3K27ac and located away from TSSs.
Chromatin accessibility + CTCF (CA–CTCF) cCREs: Accessible elements with strong CTCF binding and low histone acetylation.
Chromatin accessibility + transcription factor (CA–TF) cCREs: Accessible elements overlapping transcription factor clusters but lacking strong histone modification signals.ster.
Chromatin accessibility (CA) cCREs: Accessible elements lacking strong H3K4me3, H3K27ac, or CTCF signals.
Transcription factor (TF) cCREs: Elements defined by transcription factor binding in the absence of detectable chromatin accessibility or histone modification signals.
All signal values used for classification are retained and made available in SCREEN to support custom filtering and user-defined analyses.
Biosample-Specific Activity Annotations
In addition to cell type–agnostic classes, SCREEN annotates the biosample-specific activity of each cCRE using the corresponding epigenomic data from individual biosamples. In this context, cCREs may be annotated as active promoters, enhancers, or other classes in some biosamples and inactive in others, reflecting cell type–specific regulatory programs.
Elements lacking sufficient chromatin accessibility signal in a given biosample are labeled as having low chromatin accessibility in that context.
Core, Partial, and Ancillary Collections
Because the availability of epigenomic assays varies across biosamples, SCREEN organizes biosample-specific annotations into three collections:
- Core Collection: Biosamples with chromatin accessibility, H3K4me3, H3K27ac, and CTCF data. These provide the most complete and highest-confidence cCRE annotations and are recommended for most analyses.
- Partial Data Collection: Biosamples with chromatin accessibility and a subset of additional marks. These samples enable high-resolution boundary definition but support a reduced set of cCRE classes.
- Ancillary Collection: Biosamples lacking chromatin accessibility data. In these cases, cCREs are annotated only by the presence or absence of high signal for available assays, and users are advised to anchor analyses on cCREs defined in the Core or Partial collections.
SCREEN clearly labels biosamples by collection to guide appropriate interpretation.
Integration with Other ENCODE Annotations
SCREEN integrates cCREs with a wide range of additional ENCODE Encyclopedia annotations. For each cCRE, users can explore overlapping transcription factor binding sites, histone modification peaks, chromatin state segmentations, transcription start sites, three-dimensional chromatin interactions, and gene expression measurements. These integrations support multi-layered analyses of regulatory mechanisms.
Citation and Legacy Resources
When using cCRE annotations or analyses derived from SCREEN in publications, preprints, or other public disclosures, users should cite the appropriate corresponding publication. SCREEN provides access to and visualization of these data, but the underlying cCRE definitions, classifications, and annotations are described in the corresponding ENCODE cCRE manuscript and should be cited to ensure appropriate attribution.
Earlier versions of SCREEN remain available for reference and reproducibility but are no longer actively updated. Users should be aware that data, annotations, and features in legacy versions may differ from the current release.
Current Version:
- Registry of cCREs: Version 4
- SCREEN UI: 2026
Citation: An Expanded Registry of Candidate cis-Regulatory Elements. Moore, J.E., Pratt, H.E., Fan, K., Phalke, N., Fisher, J., Elhajjajy, S.I., Andrews, G., Gao, M., Shedd, N., Fu, Y., Lacadie, M.C., Meza, J., Khandpekar, M., Ganna, M., Choudhury, E., Swofford, R., Phan, H., Ramirez, C., Campbell, M., Likhite, M., Farrell, N.P., Weimer, A.K., Pampari, A., Ramalingam, V., Reese, F., Borsari, B., Yu, M., Wattenberg, E., Ruiz-Romero, M., Razavi-Mohseni, M., Xu, J., Galeev, T., Colubri, A., Beer, M.A., Guigó, R., Gerstein, M., Engreitz, J., Ljungman, M., Reddy, T.E., Snyder, M.P., Epstein, C.B., Gaskell, E., Bernstein, B.E., Dickel, D.E., Visel, A., Pennacchio, L.A., Mortazavi, A., Kundaje, A., Weng, Z. (2026) Nature
Legacy versions:
| Registry of cCREs Version | UI Version | URL | Citation |
|---|---|---|---|
| V4 | 2024 | screen-v4.wenglab.org | Moore…Weng (2026) Nature |
| V3 | 2020 | screen-v3.wenglab.org | |
| V2 | 2020 | screen-v2.wenglab.org | The ENCODE Project Consortium, Moore…Weng (2020) Nature |
| V1 (hg19) | 2018 | screen-v1.wenglab.org |
API Documentation
SCREEN API DocumentationContact Us
Send us a message and we'll be in touch!
As this is a beta site, we would greatly appreciate any feedback you may have. Knowing how our users are using the site and documenting issues they may have are important to make this resource better and easier to use.
If you're experiencing an error/bug, feel free to
submit an issue on Github.If you would like to send an attachment, feel free to email us directly at
encode‑screen@googlegroups.com